Submitted by Agnieszka Iwasi... on Wed, 20/04/2016 - 21:47
 Nanopore sensing is a burgeoning field with many potential applications such as DNA sequencing and protein analysis. The principle is extremely elegant: anything that passes through a tiny hole – the nanopore – filled with salty water will block it briefly. This blockage can be detected by measuring changes in ionic current driven through the nanopore by an applied potential difference. Nanopore sensing allows for the detection of the shape and charge of molecules, and is especially amenable for the analysis of biological molecules like DNA and proteins.
Nanopore sensing is a burgeoning field with many potential applications such as DNA sequencing and protein analysis. The principle is extremely elegant: anything that passes through a tiny hole – the nanopore – filled with salty water will block it briefly. This blockage can be detected by measuring changes in ionic current driven through the nanopore by an applied potential difference. Nanopore sensing allows for the detection of the shape and charge of molecules, and is especially amenable for the analysis of biological molecules like DNA and proteins.
A distinguishing feature of nanopores, in the world of biosensors, is that they are single molecule sensors meaning that they can in principle detect many sub-populations within a sample. To-date this capability of detecting sub-populations has not been fully realised for nanopore sensing since there have been no appropriate methods to simultaneously achieve selectivity and multiplexing capabilities. The possibility of multiplexing measurements of proteins has been especially difficult to achieve since proteins with similar molecular weight produce often indistinguishable signals.
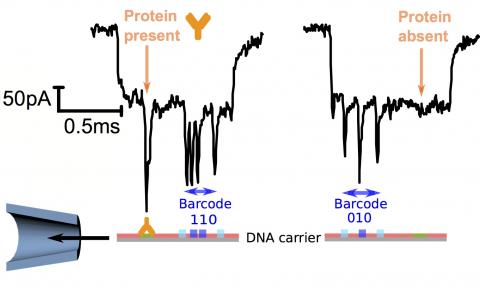
In this paper, Bell and Keyser show a remarkable new demonstration of nanopore based single molecule sensing by creating a method for accurate, selective and multiplexed analyte detection using DNA nanotechnology. Based on the programmable nature of Watson-Crick base pairing, Dr Bell designed a library of DNA structures. Each DNA structure contains a digital barcode that can be read off by the nanopore sensor by detecting tiny protrusions programmed with DNA. Each DNA barcode is constructed using a 3-bit pattern based on units of dumbbell DNA hairpins providing the necessary digital signal. This pattern could be read out with 94% accuracy from nanopore translocations. DNA conjugation techniques were then used to create tags for different proteins – namely antibodies - onto different barcodes. With this simple digital code uniquely identifying one specific antibody, the authors showed that four antibodies could be simultaneously detected from a mixture – an unprecedented result using nanopore based biosensing.
This approach provides a new avenue in protein sensing with nanopores since it simultaneously combines digitally encoded multi-analyte capabilities and selectivity. In contrast previous literature approaches of attaching a ligand to the nanopore surface target only one analyte by nature of their design. Therefore the authors believe their paper, published in the journal Nature Nanotechnology (http://dx.doi.org/10.1038/nnano.2016.50), represents an important stride for the utility of nanopore sensing for protein analysis. The authors are now investigating potential applications including making a portable system for rapid disease biomarker detection.
